Software for lattice light-sheet imaging of FRET biosensors, illustrated with a new Rap1 biosensor, J Cell Biol (2019) 218 (9): 3153–3160. DOI: 10.1083/jcb.201903019
David Adalsteinsson, the founder and CTO of Visual Data Tools is also a faculty member in the Mathematics Departement at UNC-Chapel Hill. This paper is a collaboration with Klaus Hahn in Pharmacology at UNC-Chapel Hill and the imaging group of Eric Betzig at Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA.
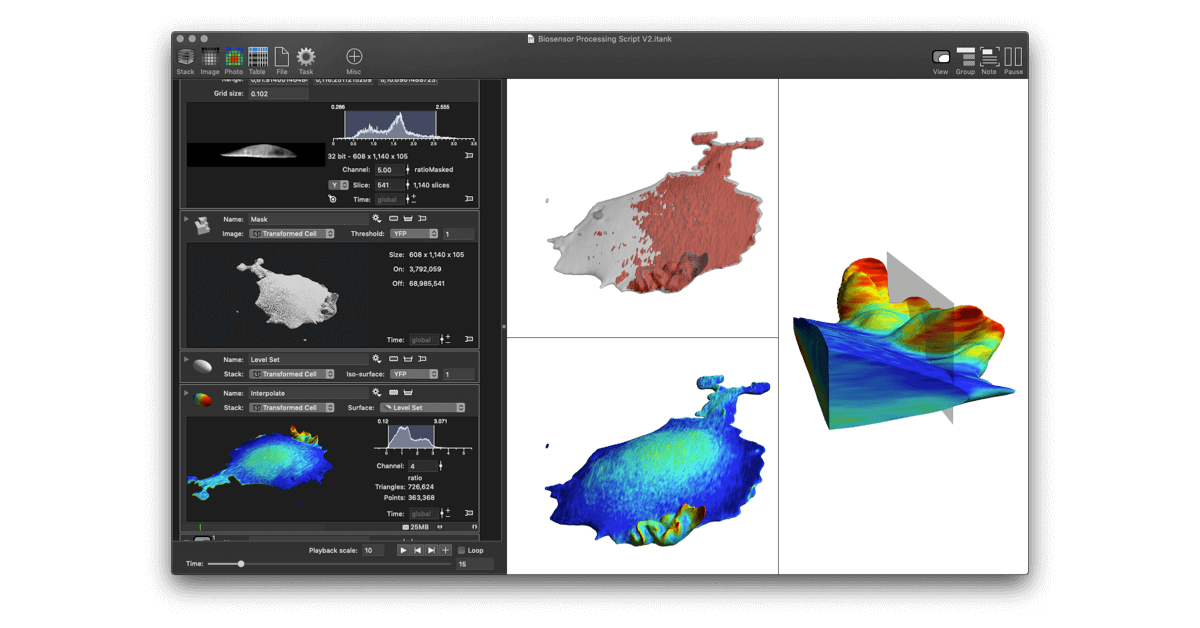
The paper demonstrates how to work with large data sets coming from Lattice Light Sheet Microscopes (LLSM) and perform manipulations needed to analyze biosensor experiments. Traditional methods using standard tools (i.e., Python, Matlab and even existing high end imaging tools) came up short in several ways. The initial work was done using DataTank, a numerical programming environment featuring 3D graphics. In order to further streamline the analysis for ease of use, a new program, ImageTank, was created. It shares the numerical foundations and code with DataTank, but uses graphical and user interface design elements from DataGraph, an application for graphing and data analysis. All three tools are distributed by Visual Data Tools, Inc.
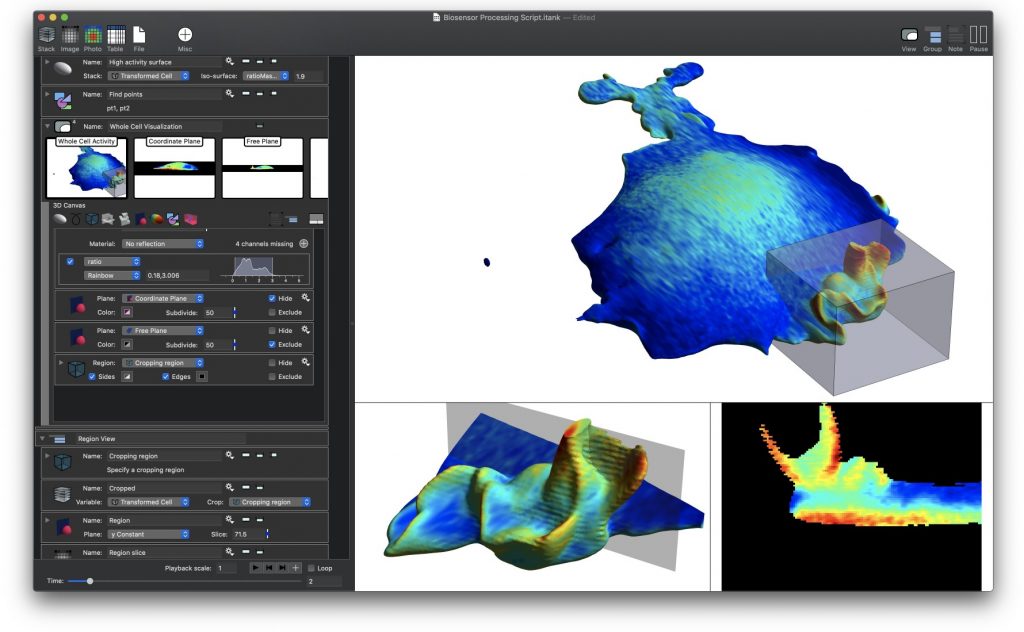
ImageTank is intended as a general purposes imaging, post-processing and scientific computing platform. It already has a number of common actions used in image processing, and is not limited to the techniques used in this paper.
ImageTank is not an open source project. It is an example of Agile Software Development, just like DataGraph. This community web site is the home for this program and the place where users can participate by asking questions, suggest features and report any issues with the program. The program is under constant development. By joining the community, you will be notified of all updates and changes to the program. See the knowledge base for information about how to install and activate the application.
There is no charge for ImageTank at this time. You can install it on any number of machines. As the program is further developed, we anticipate using an annual licensing model.
As is our goal with all the software licensed by Visual Data Tools, we anticipate charging a reasonable price and ensuring all users have access to the latest version.
Authors of the Paper
Authors: Ellen C. O’Shaughnessy, Orrin J. Stone, Paul K.LaFosse, Mihai L. Azoitei, Denis Tsygankov, John M. Heddleston, Wesley R. Legant, Erika S. Wittchen, Keith Burridge, Timothy C. Elston, Eric Betzig, Teng-Leong Chew, David Adalsteinsson, Klaus M. Hahn
Abstract
Lattice light-sheet microscopy (LLSM) is valuable for its combination of reduced photobleaching and outstanding spatiotemporal resolution in 3D. Using LLSM to image biosensors in living cells could provide unprecedented visualization of rapid, localized changes in protein conformation or posttranslational modification. However, computational manipulations required for biosensor imaging with LLSM are challenging for many software packages. The calculations require processing large amounts of data even for simple changes such as reorientation of cell renderings or testing the effects of user-selectable settings, and lattice imaging poses unique challenges in thresholding and ratio imaging. We describe here a new software package, named ImageTank, that is specifically designed for practical imaging of biosensors using LLSM. To demonstrate its capabilities, we use a new biosensor to study the rapid 3D dynamics of the small GTPase Rap1 in vesicles and cell protrusions.
Download
Click here to download the ImageTank file to run the analysis, movies that explain the process and a reduced data file (5.4GB). Click here to download the full dataset (7.2GB, compressed). Download ImageTank from here.